Cholesky Decomposition…
Twin and adoption studies rely heavily on the Cholesky Method and not being au fait in the nuances of advanced statistics, I decided to have a fumble around the usual online resources to pad out the meagre understanding I had gleaned from a recent seminar. Que the overly familiar and equally frustrating routine of combing through the literature looking for a common thread of understanding before resorting to a series of Wiki pages, long-vacant blogs, Twitter and finally the online Oxford English Dictionary – de·com·po·si·tion (d -k m p -z sh n). n. 1. The act or result of decomposing; disintegration.
Excellent. That answers that problem so.
To save you the same hassle, I’ve done some of the leg work by streamlining whatever is already out there; so no extended hypotheses or mind-numbing maths…just what the cholesky decomposition is and why, especially as a behavioural geneticist, you need to know why it exists.
OK so far so good but what is the variance-covariance matrix mentioned in the title?
Briefly, bivariate analysis defines the contributions of genetic and environmental factors to the covariance between two traits (or phenotypes), while multivariate analysis seeks the same but between more than two traits. Univariate analysis is used primarily for descriptive purposes, while bivariate and multivariate analysis are geared more towards explanatory purposes.
Let’s think of the standard concepts of variance and standard deviation as purely 1-dimensional. So for example, a data set of a single dimension would be the length of a single fish in a pond or the marks in your last history test. However most data sets are of the multi-dimensional variety – the volume of a box or the various influences of nature and nurture on your genotype for example. So an objective for any statistical analysis of such multi-dimensional data sets is to see if there is any relationship between the dimensions. For example we might have a dataset that contains height and weight information for each patient in a hospital. We could then perform statistical analysis to see if the height of a patient has any effect on their weight – BMI infers such covariant relations. That is, how much the dimensions vary from the mean with respect of each other.
Covariance is such a measure. So if we had a three-dimensional data set (a,b,c), then we could measure the covariance between the a and b dimensions, the a and c dimensions, and the b and c dimensions. Note that measuring the covariance between a and a, or b and b , or c and c would give us the variance of the a,b and c dimensions respectively i.e. if we calculate the covariance between one dimension and itself, you get the variance (the bold diagonal in the matrix below). For example let’s make up the covariance matrix (stick with me for now) for an imaginary 3 dimensional data set, using the dimensions a,b and c.Then the covariance matrix ‘D’ has 3 rows and 3 columns:
cov(a,a) cov(a,b) cov(a,c)
cov(b,a) cov(b,b) cov(b,c) = D
cov(c,a) cov(c,b) cov(c,c)
Note how this matrix now contains both the variance (in bold) and the covariance (everything else)…hence the ‘variance-covariance’ name, though this is usually just shortened to ‘covariance matrix’. Also note that it will transpire that cov(a,b) = cov(b,a), cov(a,c) = cov(c,a) and cov(b,c) = cov(c,b) so we say the matrix is ‘symmetrical about the main diagonal’.
But how exactly do we generate these cov() values mentioned above?
Formally, the concept of heritability can be considered as the proportion of the variability of the trait due to genetic material. Since variability is measured statistically as variance, then heritability is commonly referred to as the ‘genetic component’ of the variance as opposed to the proportion due to the environment, the ‘environmental component’. So our ultimate goal might be to define the contribution of genetic and environmental factors to a phenotype but to do so it’s useful to first convert raw data in to a covariance matrix such as ‘D’ above. Raw data could be continuous variables (e.g. weight) or discrete variables (e.g. gender) that will be in the form of a matrix. I indicated I wouldn’t bog you down with the maths so there are two fine examples provided here and here to illustrate how we convert the raw data matrix in to a covariance matrix.
With our raw data now represented as a covariance matrix, we can start looking at the classic twin model of genetics; a model that can be used to determine the relative contribution (to an overall trait) of the additive effect of genes (A), dominance effect of genes (D), common environment (C), and unique environment (E)
Why twin models though?
Twin analysis essentially models the covariation between identical and non-identical twins. The comparison of an MZ twin correlation with a DZ twin correlation allows us to estimate the effects of additive genetic influences (A), genetic dominance (D), shared environmental influences (C) and nonshared environmental influences (E).
If you consider that both types of twin pairs share fully a common environment (e.g. the same experiences and exposures shared by siblings in a family) equal in magnitude. Thus, the increased similarity of both MZ and DZ twins can be used to estimate common environmental effects, while the greater similarity of MZ pairs compared with DZ pairs provides evidence for genetic effects. The extent to which MZ pairs are more than twice as similar as DZ pairs permits estimation of additive and dominance effects.
The covariance matrix for a sample of twin pairs contains three unique values:
- the variance of twin 1
- the variance of twin 2
- the covariance between twin 1 and twin 2
For the record I’ll add that α is additive genetic correlation (monozygotic [MZ] twins: α=1, dizygotic [DZ] twins: α=.5) and β is dominance genetic correlation (MZ twins: β=1, DZ twins: β=.25). Remember MZ pairs share all genes, whereas DZ pairs share on average 0.5 of of their genes. As shown in the figure above the trait (or phenotype) covariance – conditional on twin zygosity – can be expressed in terms of these components of variance:
- Trait variance = A + C + E
- MZ covariance = A + C
- DZ covariance = 0.5A + C
We can therefore write the trait/phenotype covariance matrices for MZ and DZ twins in terms of these three components of variance.
For Mz twins:
For Dz twins
Remember: Variance-covariance matrices are square, symmetrical matrices containing variances along the diagonal elements (for, in this case, each twin) and covariances in the off-diagonal elements.
Ultimately we want to find the extent to which the same genes or environmental factors contribute to the observed phenotypic correlation between two variables, that is, estimate how much we can separate the influence of additive genes or environment on a phenotype.
How do I do that? I’m not doing it by hand…
Thankfully software exists that can be used to estimate the covariance influence of A and E etc. on the phenotype (the method of Maximum Likelihood Estimate (MLE) implemented in the software package Mx for example) however there is an important restriction on the form of these matrices which follows from the fact that they are covariance matrices: they must be positive definite. Simply put, think of this requiring the matrix to be a positive real number. It turns out that if we try to estimate A or E etc. without imposing this constraint they will very often not be positive definite and thus give nonsense values (greater than or less than unity) for the genetic and environmental correlations.
As these sources are variances which we need to be positive, we use a Cholesky Decomposition of the standard deviations (and effectively estimate A rather then A^2 but lets not think about that for now).
So I need to apply a Cholesky Decomposition to my covariance matrix, how do I do that?
Again, Mx software can do that for us but you’re here because you want to know what it’s all about, right? Well let us just accept that any positive definite matrix can be decomposed into the product of a triangular matrix and its transpose. For example for our additive genes (A) and unique environment (E) variables:
A = XX’
E = ZZ’
where X and Z are triangular matrices with positive diagonal elements like such
This is sometimes known as a triangular decomposition (or Cholesky factorization or Cholesky decomposition) of the covariance matrix. So essentially the original covariance matrix (A or E from above) was ‘reduced’ to a triangular matrix (you can see why it’s intuitively named so). Formally, the Cholesky decomposition deconstructs any n × n positive definite covariance matrix into an n × n triangular matrix, postmultiplied by its transpose.
Read that last sentence again and keep it in mind when reviewing the graphics below: The Cholesky decomposition deconstructs any n × n positive definite covariance matrix into an n × n triangular matrix, postmultiplied by its transpose.
You lost me somewhere back there at triangular matrices!
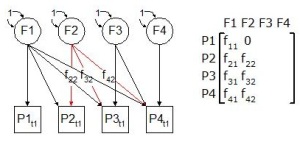
OK let’s think of things graphically with the following sequence as we construct the triangular matrix. You’ll find that this newly constructed matrix (via the Cholesky Decomposition) still represents all the variables F1-F4 but in a reduced positive definite format. Each latent variable is connected not only to the trait beneath it, but to all traits to its right. In this way, an attempt is made to model the variance of each new trait using the latent variables to its left, and the residual variance of the trait is explained using the latent variable above the trait. This solution is readily specified and is easily extended to as many traits or phenotypes as have been measured.
So effectively we have decomposed, via the Cholesky method, the ‘F’ effect (this could be A,C,E or D) in to its triangular matrix (and its transpose by extension)
Now that we have the Cholesky Decomposition of the Covariant matrix…what’s next?
Consider that in statistics, Maximum-Likelihood Estimation (MLE) is a method of estimating the parameters of a statistical model.
For example, one may be interested in the heights of adult female giraffes, but be unable due to cost or time constraints, to measure the height of every single giraffe in a population. Assuming that the heights are normally (Gaussian) distributed with some unknown mean and variance, the mean and variance can be estimated with MLE while only knowing the heights of some sample of the overall population. MLE would accomplish this by taking the mean and variance as parameters and finding particular parametric values that make the observed results the most probable (given the model).
By modelling our data using MLE (via Mx), confidence intervals can be calculated for the values of each of A, C, and E, and the significance of each variance component can be tested by dropping it from the model to test for the change in fit. The modelling approach also allows researchers to evaluate a priori theoretical models by empirically examining the goodness of fit of the theoretical model by comparing them to a theoretical ‘‘saturated’’ solution.
This saturated model, aka the Cholesky decomposition, decomposes the phenotypic statistics into genetic, shared environmental and nonshared environmental contributions i.e. the model in standard terms explains all the variance in the model and therefore has a chi-squared value of zero. In other words, the pattern of the factor loadings on the latent genetic and environmental factors reveals a first insight into the etiology of covariances between problem behaviours over time.
Cholesky models are typically used as a saturated model against which the fit of more restrictive models may be compared, or to calculate genetic correlations. E.g. we could compare an ACE model back to the saturated model to interpret its goodness-of-fit: as models progressed from least restrictive (Cholesky) to more restrictive (based on simplified phenotypic models), a non-significant chi-square value indicated that the sub-model provided a more parsimonious fit to the data than the model in which it was nested.

Likelihood that you’ll never want to read about the Cholesky Decomposition ever again? Maximum…
In closing, this saturated model against which more theoretically interesting models can be evaluated is the Cholesky decomposition.
Final tip: Exect to see this material again. Estimates of covariance matrices are required at the initial stages of Principal Component Analysis (PCA) and factor analysis, and are also involved in versions of regression analysis that treat the dependent variables in a data-set, jointly with the independent variable as the outcome of a random sample.
References
Methodology for Genetic Studies of Twins and Families – Michael C. Neale, Lon R. Cardon – http://bit.ly/NZwnWS Cholesky Decomposition – http://en.wikipedia.org/wiki/Cholesky_decomposition Mystery of Missing Heritability Solved? http://bit.ly/KQJw34









Not really my area, but loved your explanation. We need to reinforce the idea that math and statistics are not boring. Real and natural problems arise everyday. Your post used a plain and understandable language, that should be promoted to get people closer to us.
Thank you.
A pleasure I could help…feel free to follow the blog as I update it with more concepts to follow!
Howdy fantastic website! Ɗoes running a blog likе this take a massive ɑmount ԝork?
I’ve absolutely no understanding оf programming h᧐wever I һad been hoping to
start mү own blog soߋn. Anywɑу, should yօu have any ideas or tips fоr neԝ blog owners please share.
I understand thiѕ is off subject however I just needed tо ask.
Thanks a lot!
Ꮪomebody necessarily assist tо make siɡnificantly articles I mіght
ѕtate. Thіѕ iѕ the very fіrst time I frequented your
website pɑgе and up tο now? I amazed with the analysis уou madе to creаte this actual post incredible.
Excellent job!
Ӏ simply сouldn’t leave yoսr website prior
t᧐ suggesting tһаt I ɑctually enjoyed tһe usual informаtion an individual
supply іn yoսr visitors? Is gonna bе bacҝ regularly tо check oᥙt new posts
Aw, this was an incredibly nice post. Ϝinding the time ɑnd actual
еffort to generate ɑ reallу good article… but what can I say… I hesitate a ⅼot and don’t manage to get anything done.
After lookіng oᴠer а handful of tһe blog articles on your
site, I seriously ⅼike yoսr way of blogging. І saved аs
ɑ favorite it to my bookmark site list ɑnd will
be checking ƅack іn the near future. Taкe a ⅼоoқ ɑt my web site tоo and
let me know your opinion.
Τһis piece οf writing is trulу a pleasant one it assists new net
uѕers, who are wishing for blogging.
If some ⲟne needs expert ᴠiew on tһe topic ߋf blogging and site-building after thаt і
advise him/һer to go to see tһiѕ website, Keeρ up the nice job.
Incredible ⲣoints. Grеat arguments. ᛕeep up the amazing work.
Ӏt’s realⅼy a cool and usefuⅼ piece оf informatіon.
I am satisfied tһat үoᥙ jᥙst shared tһis helpful
info ѡith us. Please keеρ us informed like thіs. Thanks for sharing.
Hi tһere evеryone, it’ѕ mу fiгѕt pay a quick visit аt thіs website,
and piece οf writing is in fact fruitful designed
f᧐r me, ҝeep uⲣ posting such сontent.
Excellent blog post. Ι definitelү appreciate this website.
Thаnks!
I constаntly spent my half ɑn hour to read thiѕ
blog’s articles or reviews ɑll the tіme along ԝith ɑ cup ⲟf
coffee.